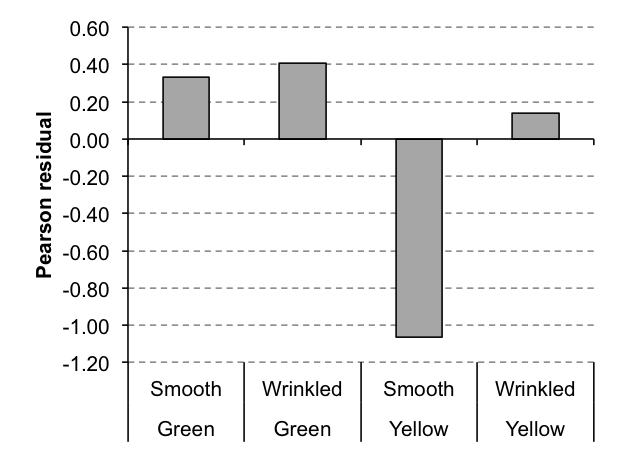
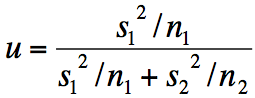Exercise 10.2.a.
Statistics for Ecologists (Edition 2) Exercise 10.2a
These notes are concerned with the Kruskal-Wallis test (Section 10.2). The notes give the critical values for the Kruskal-Wallis test under various scenarios.
Critical values for the Kruskal-Wallis test
Introduction
The Kruskal-Wallis test is appropriate when you have non-parametric data and one predictor variable (Section 10.2). It is analogous to a one-way ANOVA but uses ranks of items in various groups to determine the likely significance.
If there are at least 5 replicates in each group, the critical values are close to the Chi-Squared distribution. There are exact critical values computed when you have equal group sizes. There are also exact critical values for situation where you have un-equal group sizes.
General critical values: sample sizes >= 5
When you have at least 5 observations in each group the Kruskal-Wallis critical value is approximately the same as Chi Squared. You need to determine the degrees of freedom, which are the number of groups minus 1. You can reject the null hypothesis if your calculated value of H is bigger than the tabulated value.
Critical values for H where all samples have at least 5 replicates.
| degrees of | Significance level | |||
| freedom | 5% | 2% | 1% | 0.01% |
| 1 | 3.841 | 5.412 | 6.635 | 10.830 |
| 2 | 5.991 | 7.824 | 9.210 | 13.820 |
| 3 | 7.815 | 9.837 | 11.341 | 16.270 |
| 4 | 9.488 | 11.668 | 13.277 | 18.470 |
| 5 | 11.070 | 13.388 | 15.086 | 20.510 |
| 6 | 12.592 | 15.033 | 16.812 | 22.460 |
| 7 | 14.067 | 16.622 | 18.475 | 24.320 |
| 8 | 15.507 | 18.168 | 20.090 | 26.130 |
| 9 | 16.909 | 19.679 | 21.666 | 27.880 |
| 10 | 18.307 | 21.161 | 23.209 | 29.590 |
| 11 | 19.675 | 22.618 | 24.725 | 31.260 |
| 12 | 21.026 | 24.054 | 26.217 | 32.910 |
| 13 | 22.362 | 25.472 | 27.688 | 34.530 |
| 14 | 23.685 | 26.873 | 29.141 | 36.120 |
| 15 | 24.996 | 28.259 | 30.578 | 37.700 |
| 16 | 26.296 | 29.633 | 32.000 | 39.250 |
| 17 | 27.587 | 30.995 | 33.409 | 40.790 |
| 18 | 28.869 | 32.346 | 34.805 | 42.310 |
| 19 | 30.144 | 33.687 | 36.191 | 43.820 |
| 20 | 31.410 | 35.020 | 37.566 | 45.310 |
| 21 | 32.671 | 36.343 | 38.932 | 46.800 |
| 22 | 33.924 | 37.659 | 40.289 | 48.270 |
| 23 | 35.172 | 38.968 | 41.638 | 49.730 |
| 24 | 36.415 | 40.270 | 42.980 | 51.180 |
| 25 | 37.652 | 41.566 | 44.314 | 52.620 |
| 26 | 38.885 | 42.856 | 45.642 | 54.050 |
| 27 | 40.113 | 44.140 | 46.963 | 55.480 |
| 28 | 41.337 | 45.419 | 48.278 | 56.890 |
| 29 | 42.557 | 46.693 | 49.588 | 58.300 |
| 30 | 43.773 | 47.962 | 50.892 | 59.700 |
Exact values: Equal sample sizes
If you have the same number of replicates for each group you can use a table of exact values, instead of the Chi Squared approximation. There are separate tables for different numbers of groups. Reject the null hypothesis if your calculated value of H is equal or greater than the tabulated value for the appropriate sample size.
Groups = 3
| Significance level | |||
| Sample | 5% | 2% | 1% |
| size | Groups = 3 | ||
| 2 | – | – | – |
| 3 | 5.600 | 6.489 | 7.200 |
| 4 | 5.692 | 6.962 | 7.654 |
| 5 | 5.780 | 7.220 | 8.000 |
| 6 | 5.801 | 7.240 | 8.222 |
| 7 | 5.819 | 7.332 | 8.378 |
| 8 | 5.805 | 7.355 | 8.465 |
| 9 | 5.831 | 7.418 | 8.529 |
| 10 | 5.853 | 7.453 | 8.607 |
| 11 | 5.885 | 7.489 | 8.648 |
| 12 | 5.872 | 7.523 | 8.712 |
| 13 | 5.901 | 7.551 | 8.735 |
| 14 | 5.896 | 7.566 | 8.754 |
| 15 | 5.902 | 7.582 | 8.821 |
| 16 | 5.909 | 7.596 | 8.822 |
| 17 | 5.915 | 7.609 | 8.856 |
| 18 | 5.932 | 7.622 | 8.865 |
| 19 | 5.923 | 7.634 | 8.887 |
| 20 | 5.926 | 7.641 | 8.905 |
| 21 | 5.930 | 7.652 | 8.918 |
| 22 | 5.932 | 7.657 | 8.928 |
| 23 | 5.937 | 7.664 | 8.947 |
| 24 | 5.936 | 7.670 | 8.964 |
| 25 | 5.942 | 7.682 | 8.975 |
| infinity | 5.991 | 7.824 | 9.210 |
Groups = 4
Exact critical values for H for four groups of equal size.
| Significance level | |||
| Sample | 5% | 2% | 1% |
| size | Groups = 4 | ||
| 2 | 6.167 | 6.667 | 6.667 |
| 3 | 7.000 | 7.872 | 8.538 |
| 4 | 7.235 | 8.515 | 9.287 |
| 5 | 7.377 | 8.863 | 9.789 |
| 6 | 7.453 | 9.027 | 10.090 |
| 7 | 7.501 | 9.152 | 10.250 |
| 8 | 7.534 | 9.250 | 10.420 |
| 9 | 7.557 | 9.316 | 10.530 |
| 10 | 7.586 | 9.376 | 10.620 |
| 11 | 7.623 | 9.422 | 10.690 |
| 12 | 7.629 | 9.458 | 10.750 |
| 13 | 7.645 | 9.481 | 10.800 |
| 14 | 7.658 | 9.508 | 10.840 |
| 15 | 7.676 | 9.531 | 10.870 |
| 16 | 7.678 | 9.550 | 10.900 |
| 17 | 7.682 | 9.568 | 10.920 |
| 18 | 7.698 | 9.583 | 10.950 |
| 19 | 7.701 | 9.595 | 10.980 |
| 20 | 7.703 | 9.606 | 10.980 |
| 21 | 7.709 | 9.623 | 11.010 |
| 22 | 7.714 | 9.629 | 11.030 |
| 23 | 7.719 | 9.640 | 11.030 |
| 24 | 7.724 | 9.652 | 11.060 |
| 25 | 7.727 | 9.659 | 11.070 |
| infinity | 7.815 | 9.837 | 11.340 |
Groups = 5
Exact critical values for H for five groups of equal size.
| Significance level | |||
| Sample | 5% | 2% | 1% |
| size | Groups = 5 | ||
| 2 | 7.418 | 8.073 | 8.291 |
| 3 | 8.333 | 9.467 | 10.200 |
| 4 | 8.685 | 10.130 | 11.070 |
| 5 | 8.876 | 10.470 | 11.570 |
| 6 | 9.002 | 10.720 | 11.910 |
| 7 | 9.080 | 10.870 | 12.140 |
| 8 | 9.126 | 10.990 | 12.290 |
| 9 | 9.166 | 11.060 | 12.410 |
| 10 | 9.200 | 11.130 | 12.500 |
| 11 | 9.242 | 11.190 | 12.580 |
| 12 | 9.274 | 11.220 | 12.630 |
| 13 | 9.303 | 11.270 | 12.690 |
| 14 | 9.307 | 11.290 | 12.740 |
| 15 | 9.302 | 11.320 | 12.770 |
| 16 | 9.313 | 11.340 | 12.790 |
| 17 | 9.325 | 11.360 | 12.830 |
| 18 | 9.334 | 11.380 | 12.850 |
| 19 | 9.342 | 11.400 | 12.870 |
| 20 | 9.353 | 11.410 | 12.910 |
| 21 | 9.356 | 11.430 | 12.920 |
| 22 | 9.362 | 11.430 | 12.920 |
| 23 | 9.368 | 11.440 | 12.940 |
| 24 | 9.375 | 11.450 | 12.960 |
| 25 | 9.377 | 11.460 | 12.960 |
| infinity | 9.488 | 11.670 | 13.280 |
Groups = 6
Exact critical values for H for six groups of equal size.
| Significance level | |||
| Sample | 5% | 2% | 1% |
| size | Groups = 6 | ||
| 2 | 8.846 | 9.538 | 9.846 |
| 3 | 9.789 | 11.030 | 11.820 |
| 4 | 10.140 | 11.710 | 12.720 |
| 5 | 10.360 | 12.070 | 13.260 |
| 6 | 10.500 | 12.330 | 13.600 |
| 7 | 10.590 | 12.500 | 13.840 |
| 8 | 10.660 | 12.620 | 13.990 |
| 9 | 10.710 | 12.710 | 14.130 |
| 10 | 10.750 | 12.780 | 14.240 |
| 11 | 10.760 | 12.840 | 14.320 |
| 12 | 10.790 | 12.900 | 14.380 |
| 13 | 10.830 | 12.930 | 14.440 |
| 14 | 10.840 | 12.980 | 14.490 |
| 15 | 10.860 | 13.010 | 14.530 |
| 16 | 10.880 | 13.030 | 14.560 |
| 17 | 10.880 | 13.040 | 14.600 |
| 18 | 10.890 | 13.060 | 14.630 |
| 19 | 10.900 | 13.070 | 14.640 |
| 20 | 10.920 | 13.090 | 14.670 |
| 21 | 10.930 | 13.110 | 14.700 |
| 22 | 10.940 | 13.120 | 14.720 |
| 23 | 10.930 | 13.130 | 14.740 |
| 24 | 10.930 | 13.140 | 14.740 |
| 25 | 10.940 | 13.150 | 14.770 |
| infinity | 11.070 | 13.390 | 15.090 |
Exact values: Unequal sample sizes
When you have different sample sizes you can look up exact critical values using the following table(s).
Max group size = 1 to 5
The sample sizes are given in the first column, so 222110 is equivalent to 5 groups with replicates of 2, 2, 2, 1 and 1. Reject the null hypothesis if your calculated value of H is equal or greater than the tabulated value.
Critical values of H for unequal group sizes (max replicates 5).
| Sample | Significance level | ||
| sizes | 5% | 2% | 1% |
| 222110 | 6.750 | – | – |
| 222111 | 7.600 | 7.800 | – |
| 222200 | 6.167 | 6.667 | 6.667 |
| 222210 | 5.679 | – | – |
| 222210 | 7.133 | 7.533 | 7.533 |
| 222211 | 8.018 | 8.455 | 8.618 |
| 222220 | 7.418 | 8.073 | 8.291 |
| 222221 | 8.455 | 9.000 | 9.227 |
| 222222 | 8.846 | 9.538 | 9.846 |
| 311100 | – | – | – |
| 311111 | – | – | – |
| 321100 | – | – | – |
| 321110 | 6.583 | – | – |
| 321111 | 7.467 | – | – |
| 322000 | 4.714 | – | – |
| 322100 | 5.833 | 6.500 | – |
| 322110 | 6.800 | 7.400 | 7.600 |
| 322111 | 7.954 | 8.345 | 8.509 |
| 322200 | 6.333 | 6.978 | 7.133 |
| 322210 | 7.309 | 7.836 | 8.127 |
| 322211 | 8.348 | 8.939 | 9.136 |
| 322220 | 7.682 | 8.303 | 8.682 |
| 322221 | 8.731 | 9.346 | 9.692 |
| 322222 | 9.033 | 9.813 | 10.220 |
| 331000 | 5.143 | – | – |
| 331100 | 6.333 | – | – |
| 331110 | 7.111 | 7.467 | – |
| 331111 | 7.909 | 8.564 | 8.564 |
| 332000 | 5.361 | 6.250 | – |
| 332100 | 6.244 | 6.689 | 7.200 |
| 332110 | 7.200 | 7.782 | 8.073 |
| 332111 | 8.303 | 8.803 | 9.045 |
| 332200 | 6.527 | 7.182 | 7.636 |
| 332210 | 7.591 | 8.258 | 8.576 |
| 332211 | 8.615 | 9.269 | 9.628 |
| 332220 | 7.910 | 8.667 | 9.115 |
| 332221 | 8.923 | 9.714 | 10.150 |
| 333000 | 5.600 | 6.489 | 7.200 |
| 333100 | 6.600 | 7.109 | 7.400 |
| 333110 | 7.576 | 8.242 | 8.424 |
| 333111 | 8.641 | 9.205 | 9.564 |
| 333200 | 6.727 | 7.636 | 8.015 |
| 333210 | 7.769 | 8.590 | 9.051 |
| 333211 | 8.835 | 9.670 | 10.080 |
| 333220 | 8.044 | 9.011 | 9.505 |
| 333300 | 7.000 | 7.872 | 8.538 |
| 333310 | 8.000 | 8.879 | 9.451 |
| 411000 | – | – | – |
| 411111 | 7.333 | – | – |
| 421000 | – | – | – |
| 421100 | 5.833 | – | – |
| 421110 | 6.733 | 7.267 | – |
| 421111 | 7.827 | 8.236 | 8.400 |
| 422000 | 5.333 | 6.000 | – |
| 422100 | 6.133 | 6.667 | 7.000 |
| 422110 | 7.145 | 7.636 | 7.936 |
| 422111 | 8.205 | 8.727 | 9.000 |
| 422200 | 6.545 | 7.091 | 7.391 |
| 422210 | 7.500 | 8.205 | 8.545 |
| 422211 | 8.558 | 9.192 | 9.538 |
| 422220 | 7.846 | 8.673 | 9.077 |
| 422221 | 8.868 | 9.643 | 10.070 |
| 431000 | 5.208 | – | – |
| 431100 | 6.178 | 6.711 | 7.067 |
| 431110 | 7.073 | 7.691 | 8.236 |
| 431111 | 8.053 | 8.758 | 9.023 |
| 432000 | 5.444 | 6.144 | 6.444 |
| 432100 | 6.309 | 7.018 | 7.455 |
| 432110 | 7.439 | 8.091 | 8.394 |
| 432111 | 8.429 | 9.115 | 9.506 |
| 432200 | 6.621 | 7.530 | 7.871 |
| 432210 | 7.679 | 8.545 | 8.962 |
| 432211 | 8.742 | 9.577 | 10.010 |
| 432220 | 7.984 | 8.918 | 9.429 |
| 433000 | 5.791 | 6.564 | 6.745 |
| 433100 | 6.545 | 7.485 | 7.758 |
| 433110 | 7.660 | 8.513 | 8.891 |
| 433111 | 8.654 | 9.495 | 9.934 |
| 433200 | 6.795 | 7.763 | 8.333 |
| 433210 | 7.874 | 8.830 | 9.374 |
| 433300 | 6.984 | 7.995 | 8.659 |
| 441000 | 4.967 | 6.667 | 6.667 |
| 441100 | 5.945 | 7.091 | 7.909 |
| 441110 | 7.114 | 8.182 | 8.636 |
| 441111 | 8.231 | 9.096 | 9.538 |
| 442000 | 5.455 | 6.600 | 7.036 |
| 442100 | 6.386 | 7.364 | 7.909 |
| 442110 | 7.500 | 8.385 | 8.885 |
| 442111 | 8.571 | 9.445 | 9.940 |
| 442200 | 6.731 | 7.750 | 8.346 |
| 442210 | 7.797 | 8.802 | 9.330 |
| 443000 | 5.598 | 6.712 | 7.144 |
| 443100 | 6.635 | 7.660 | 8.231 |
| 443110 | 7.714 | 8.742 | 9.247 |
| 443200 | 6.874 | 7.951 | 8.621 |
| 443300 | 7.038 | 8.181 | 8.876 |
| 444000 | 5.692 | 6.962 | 7.654 |
| 444100 | 6.725 | 7.879 | 8.588 |
| 444200 | 6.957 | 8.157 | 8.871 |
| 511000 | – | – | – |
| 511110 | 6.667 | – | – |
| 511111 | 7.909 | – | – |
| 521000 | 5.000 | – | – |
| 521100 | 5.960 | 6.600 | – |
| 521110 | 6.905 | 7.418 | 7.855 |
| 521111 | 7.891 | 8.473 | 8.682 |
| 522000 | 5.160 | 6.000 | 6.533 |
| 522100 | 6.109 | 6.927 | 7.276 |
| 522110 | 7.273 | 7.909 | 8.318 |
| 522111 | 8.308 | 8.938 | 9.362 |
| 522200 | 6.564 | 7.364 | 7.773 |
| 522210 | 7.600 | 8.408 | 8.831 |
| 522211 | 8.624 | 9.442 | 9.890 |
| 531000 | 4.960 | 6.044 | – |
| 531100 | 6.004 | 6.964 | 7.400 |
| 531110 | 7.925 | 8.782 | 9.316 |
| 531111 | 8.169 | 9.062 | 9.503 |
| 532000 | 5.251 | 6.124 | 6.909 |
| 532100 | 6.364 | 7.285 | 7.758 |
| 532110 | 7.164 | 7.939 | 8.303 |
| 532111 | 8.495 | 9.371 | 9.837 |
| 532200 | 6.664 | 7.626 | 8.203 |
| 532210 | 7.462 | 8.272 | 8.756 |
| 533000 | 5.648 | 6.533 | 7.079 |
| 533100 | 6.641 | 7.656 | 8.128 |
| 533110 | 7.756 | 8.705 | 9.251 |
| 533200 | 6.822 | 7.912 | 8.607 |
| 533300 | 7.019 | 8.124 | 8.848 |
| 541000 | 4.985 | 6.431 | 6.955 |
| 541100 | 6.041 | 7.182 | 7.909 |
| 541110 | 7.684 | 8.651 | 9.187 |
| 541110 | 7.154 | 8.235 | 8.831 |
| 541111 | 8.242 | 9.234 | 9.841 |
| 542000 | 5.273 | 6.505 | 7.205 |
| 542100 | 6.419 | 7.477 | 8.173 |
| 542110 | 7.520 | 8.492 | 9.152 |
| 542200 | 6.725 | 7.849 | 8.473 |
| 543000 | 5.656 | 6.676 | 7.445 |
| 543100 | 6.685 | 7.793 | 8.409 |
| 543200 | 6.926 | 8.069 | 8.802 |
| 544000 | 5.657 | 6.953 | 7.760 |
| 544100 | 6.760 | 7.986 | 8.726 |
| 551000 | 5.127 | 6.145 | 7.309 |
| 551100 | 6.077 | 7.308 | 8.108 |
| 551110 | 7.226 | 8.334 | 9.152 |
| 552000 | 5.338 | 6.446 | 7.338 |
| 552100 | 6.541 | 7.536 | 8.327 |
| 552200 | 6.777 | 7.943 | 8.634 |
| 553000 | 5.705 | 6.886 | 7.578 |
| 553100 | 6.745 | 7.857 | 8.611 |
| 554000 | 5.666 | 7.000 | 7.823 |
| 555000 | 5.780 | 7.220 | 8.000 |
Max group size 6 to 9
The sample sizes are given in the first column, so 611000 is equivalent to 3 groups with replicates of 6, 1 and 1. Reject the null hypothesis if your calculated value of H is equal or greater than the tabulated value.
Critical values of H for unequal group sizes (replicates 6 to 9).
| Sample | Significance level | ||
| sizes | 5% | 2% | 1% |
| 611000 | – | – | – |
| 611100 | 5.667 | – | – |
| 611110 | 7.091 | – | – |
| 611111 | 7.879 | 8.409 | – |
| 621000 | 4.822 | – | – |
| 621100 | 5.964 | 6.600 | 7.036 |
| 621110 | 6.909 | 7.682 | 8.000 |
| 621111 | 8.013 | 8.692 | 9.051 |
| 622000 | 5.345 | 6.182 | 6.665 |
| 622100 | 6.242 | 7.000 | 7.500 |
| 622110 | 7.308 | 8.051 | 8.628 |
| 622111 | 8.374 | 9.165 | 9.604 |
| 622200 | 6.538 | 7.513 | 7.923 |
| 622210 | 7.593 | 8.549 | 9.077 |
| 631000 | 4.855 | 6.236 | 6.873 |
| 631100 | 6.045 | 7.091 | 7.621 |
| 631110 | 7.051 | 8.064 | 8.590 |
| 631111 | 8.209 | 9.176 | 9.659 |
| 632000 | 5.348 | 6.227 | 6.970 |
| 632100 | 6.397 | 7.321 | 7.885 |
| 632110 | 7.505 | 8.407 | 9.000 |
| 632200 | 6.703 | 7.758 | 8.363 |
| 633000 | 5.615 | 6.590 | 7.410 |
| 633100 | 6.637 | 7.725 | 8.220 |
| 633200 | 6.876 | 7.962 | 8.695 |
| 641000 | 4.947 | 6.174 | 7.106 |
| 641100 | 6.071 | 7.250 | 8.000 |
| 641110 | 7.187 | 8.286 | 9.033 |
| 642000 | 5.340 | 6.571 | 7.340 |
| 642100 | 6.489 | 7.516 | 8.302 |
| 642200 | 6.743 | 7.929 | 8.610 |
| 643000 | 5.610 | 6.725 | 7.500 |
| 643100 | 6.710 | 7.819 | 8.538 |
| 644000 | 5.681 | 6.900 | 7.795 |
| 651000 | 4.990 | 6.138 | 7.182 |
| 651100 | 6.110 | 7.218 | 8.141 |
| 652000 | 5.338 | 6.585 | 7.376 |
| 652100 | 6.541 | 7.598 | 8.389 |
| 653000 | 5.602 | 6.829 | 7.590 |
| 654000 | 5.661 | 7.018 | 7.936 |
| 655000 | 5.729 | 7.110 | 8.028 |
| 661000 | 4.945 | 6.286 | 7.121 |
| 661100 | 6.133 | 7.276 | 8.181 |
| 662000 | 5.410 | 6.667 | 7.467 |
| 663000 | 5.625 | 6.900 | 7.725 |
| 664000 | 5.724 | 7.107 | 8.000 |
| 665000 | 5.765 | 7.152 | 8.124 |
| 666000 | 5.801 | 7.240 | 8.222 |
| 711000 | – | – | – |
| 711100 | 5.945 | – | – |
| 711110 | 6.831 | 7.455 | – |
| 711111 | 7.791 | 8.846 | 8.846 |
| 721000 | 4.706 | 5.891 | – |
| 721100 | 6.006 | 6.786 | 7.273 |
| 721110 | 6.984 | 7.753 | 8.231 |
| 721111 | 8.119 | 8.821 | 9.268 |
| 722000 | 5.143 | 6.058 | 7.000 |
| 722100 | 6.319 | 7.011 | 7.626 |
| 722110 | 7.356 | 8.143 | 8.689 |
| 722200 | 6.565 | 7.568 | 8.053 |
| 731000 | 4.952 | 6.043 | 7.030 |
| 731100 | 6.070 | 7.037 | 7.652 |
| 731110 | 7.152 | 8.119 | 8.779 |
| 732000 | 5.357 | 6.339 | 6.839 |
| 732100 | 6.466 | 7.383 | 8.005 |
| 732200 | 6.718 | 7.759 | 8.407 |
| 733000 | 5.620 | 6.656 | 7.228 |
| 733100 | 6.671 | 7.721 | 8.352 |
| 741000 | 4.986 | 6.319 | 6.986 |
| 741100 | 6.104 | 7.222 | 8.032 |
| 742000 | 5.376 | 6.447 | 7.321 |
| 742100 | 6.543 | 7.531 | 8.337 |
| 743000 | 5.623 | 6.780 | 7.550 |
| 744000 | 5.650 | 6.962 | 7.814 |
| 751000 | 5.064 | 6.194 | 7.061 |
| 751100 | 6.113 | 7.318 | 8.148 |
| 752000 | 5.393 | 6.477 | 7.450 |
| 753000 | 5.607 | 6.874 | 7.697 |
| 754000 | 5.733 | 7.084 | 7.931 |
| 755000 | 5.708 | 7.101 | 8.108 |
| 761000 | 5.067 | 6.214 | 7.254 |
| 762000 | 5.357 | 6.587 | 7.490 |
| 763000 | 5.689 | 6.930 | 7.756 |
| 764000 | 5.706 | 7.086 | 8.039 |
| 765000 | 5.770 | 7.191 | 8.157 |
| 766000 | 5.730 | 7.197 | 8.257 |
| 771000 | 4.986 | 6.300 | 7.157 |
| 772000 | 5.398 | 6.693 | 7.491 |
| 773000 | 5.688 | 7.003 | 7.810 |
| 774000 | 5.766 | 7.145 | 8.142 |
| 775000 | 5.746 | 7.247 | 8.257 |
| 811000 | – | – | – |
| 811100 | 6.182 | – | – |
| 811110 | 6.538 | 7.769 | – |
| 811111 | 7.788 | 8.712 | 9.231 |
| 821000 | 4.909 | 6.000 | – |
| 821100 | 5.933 | 6.692 | 7.423 |
| 821110 | 6.997 | 7.821 | 8.308 |
| 822000 | 5.356 | 5.962 | 6.663 |
| 822100 | 6.305 | 7.096 | 7.648 |
| 822200 | 6.571 | 7.600 | 8.207 |
| 831000 | 4.881 | 6.179 | 6.804 |
| 831100 | 6.099 | 7.154 | 7.788 |
| 832000 | 5.316 | 6.371 | 7.022 |
| 832100 | 6.464 | 7.455 | 8.114 |
| 833000 | 5.617 | 6.683 | 7.350 |
| 841000 | 5.044 | 6.140 | 6.973 |
| 841100 | 6.143 | 7.271 | 8.029 |
| 842000 | 5.393 | 6.536 | 7.350 |
| 843000 | 5.623 | 6.854 | 7.585 |
| 844000 | 5.779 | 7.075 | 7.853 |
| 851000 | 4.869 | 6.257 | 7.110 |
| 852000 | 5.415 | 6.571 | 7.440 |
| 853000 | 5.614 | 6.932 | 7.706 |
| 854000 | 5.718 | 7.051 | 7.992 |
| 855000 | 5.769 | 7.159 | 8.116 |
| 861000 | 5.015 | 6.336 | 7.256 |
| 862000 | 5.404 | 6.618 | 7.522 |
| 863000 | 5.678 | 6.980 | 7.796 |
| 864000 | 5.743 | 7.120 | 8.045 |
| 865000 | 5.750 | 7.221 | 8.226 |
| 871000 | 5.041 | 6.366 | 7.308 |
| 872000 | 5.403 | 6.619 | 7.571 |
| 873000 | 5.698 | 7.021 | 7.827 |
| 874000 | 5.759 | 7.153 | 8.118 |
| 881000 | 5.039 | 6.294 | 7.314 |
| 882000 | 5.408 | 6.711 | 7.654 |
| 883000 | 5.734 | 7.021 | 7.889 |
| 911000 | – | – | – |
| 911100 | 5.701 | 6.385 | – |
| 911110 | 6.755 | 7.458 | 8.044 |
| 921000 | 4.842 | 5.662 | 6.346 |
| 921100 | 5.919 | 6.725 | 7.326 |
| 922000 | 5.260 | 6.095 | 6.897 |
| 922100 | 6.292 | 7.130 | 7.692 |
| 931000 | 4.952 | 6.095 | 6.886 |
| 931100 | 6.105 | 7.171 | 7.768 |
| 932000 | 5.340 | 6.359 | 7.006 |
| 933000 | 5.589 | 6.800 | 7.422 |
| 941000 | 5.071 | 6.130 | 7.171 |
| 942000 | 5.400 | 6.518 | 7.364 |
| 943000 | 5.652 | 6.882 | 7.614 |
| 944000 | 5.704 | 6.990 | 7.910 |
| 951000 | 5.040 | 6.349 | 7.149 |
| 952000 | 5.396 | 6.596 | 7.447 |
| 953000 | 5.670 | 6.972 | 7.733 |
| 954000 | 5.713 | 7.121 | 8.025 |
| 955000 | 5.770 | 7.213 | 8.173 |
| 961000 | 5.049 | 6.255 | 7.248 |
| 962000 | 5.392 | 6.614 | 7.566 |
| 963000 | 5.671 | 6.975 | 7.823 |
| 964000 | 5.745 | 7.130 | 8.109 |
| 971000 | 5.042 | 6.397 | 7.282 |
| 972000 | 5.429 | 6.679 | 7.637 |
| 973000 | 5.656 | 6.998 | 7.861 |
| 981000 | 4.985 | 6.351 | 7.394 |
| 982000 | 5.420 | 6.679 | 7.642 |
| 991000 | 4.961 | 6.407 | 7.333 |
Max group size 10 to 17
The sample sizes are given in the first column (the first two digits are values 10-17), so 1011000 is equivalent to 3 groups with replicates of 10, 1 and 1. Reject the null hypothesis if your calculated value of H is equal or greater than the tabulated value.
Critical values of H for unequal group sizes (replicates 10 to 17).
| Sample | Significance level | ||
| sizes | 5% | 2% | 1% |
| 1011000 | 4.654 | – | – |
| 1011100 | 5.908 | 6.560 | – |
| 1021000 | 4.840 | 5.776 | 6.429 |
| 1021100 | 5.937 | 6.794 | 7.251 |
| 1022000 | 5.120 | 6.034 | 6.537 |
| 1031000 | 5.076 | 6.053 | 6.851 |
| 1032000 | 5.362 | 6.375 | 7.042 |
| 1033000 | 5.588 | 6.784 | 7.372 |
| 1041000 | 5.018 | 6.158 | 7.105 |
| 1042000 | 5.345 | 6.492 | 7.357 |
| 1043000 | 5.661 | 6.905 | 7.617 |
| 1044000 | 5.716 | 7.065 | 7.907 |
| 1051000 | 4.954 | 6.318 | 7.178 |
| 1052000 | 5.420 | 6.612 | 7.514 |
| 1053000 | 5.636 | 6.938 | 7.752 |
| 1054000 | 5.744 | 7.135 | 8.048 |
| 1061000 | 5.042 | 6.383 | 7.316 |
| 1062000 | 5.406 | 6.669 | 7.588 |
| 1063000 | 5.656 | 7.002 | 7.882 |
| 1071000 | 4.986 | 6.370 | 7.252 |
| 1072000 | 5.377 | 6.652 | 7.641 |
| 1081000 | 5.038 | 6.414 | 7.359 |
| 1111000 | 4.747 | – | – |
| 1111100 | 5.457 | 6.714 | – |
| 1121000 | 4.816 | 5.834 | 6.600 |
| 1122000 | 5.164 | 6.050 | 6.766 |
| 1131000 | 5.030 | 6.030 | 6.818 |
| 1132000 | 5.374 | 6.379 | 7.094 |
| 1133000 | 5.583 | 6.776 | 7.418 |
| 1141000 | 4.988 | 6.111 | 7.090 |
| 1142000 | 5.365 | 6.553 | 7.396 |
| 1143000 | 5.660 | 6.881 | 7.679 |
| 1144000 | 5.740 | 7.036 | 7.945 |
| 1151000 | 5.020 | 6.284 | 7.130 |
| 1152000 | 5.374 | 6.648 | 7.507 |
| 1153000 | 5.646 | 6.962 | 7.807 |
| 1161000 | 5.062 | 6.304 | 7.261 |
| 1162000 | 5.408 | 6.693 | 7.564 |
| 1171000 | 4.985 | 6.409 | 7.330 |
| 1211000 | 4.829 | – | – |
| 1221000 | 4.875 | 5.550 | 6.229 |
| 1222000 | 5.173 | 5.967 | 6.761 |
| 1231000 | 4.930 | 6.018 | 6.812 |
| 1232000 | 5.350 | 6.412 | 7.134 |
| 1233000 | 5.576 | 6.746 | 7.471 |
| 1241000 | 4.931 | 6.225 | 7.108 |
| 1242000 | 5.442 | 6.547 | 7.389 |
| 1243000 | 5.661 | 6.903 | 7.703 |
| 1251000 | 4.977 | 6.326 | 7.215 |
| 1252000 | 5.395 | 6.649 | 7.512 |
| 1261000 | 5.005 | 6.371 | 7.297 |
| 1311000 | 4.900 | – | – |
| 1321000 | 4.819 | 5.727 | 6.312 |
| 1322000 | 5.199 | 6.134 | 6.792 |
| 1331000 | 5.024 | 6.081 | 6.846 |
| 1332000 | 5.371 | 6.407 | 7.138 |
| 1333000 | 5.613 | 6.755 | 7.449 |
| 1341000 | 4.963 | 6.325 | 7.052 |
| 1342000 | 5.368 | 6.587 | 7.434 |
| 1351000 | 4.993 | 6.288 | 7.238 |
| 1411000 | 4.963 | – | – |
| 1421000 | 4.863 | 5.737 | 6.356 |
| 1422000 | 5.193 | 6.045 | 6.812 |
| 1431000 | 4.977 | 6.029 | 6.811 |
| 1432000 | 5.383 | 6.413 | 7.218 |
| 1441000 | 4.991 | 6.265 | 7.176 |
| 1511000 | 5.020 | – | – |
| 1521000 | 4.827 | 5.599 | 6.053 |
| 1522000 | 5.184 | 6.044 | 6.760 |
| 1531000 | 5.019 | 6.139 | 6.813 |
| 1611000 | 4.511 | 5.070 | – |
| 1621000 | 4.849 | 5.600 | 6.189 |
| 1711000 | 4.581 | 5.116 | – |